--- title: "GoMex cetacean & sea turtle SDMs" editor_options: chunk_output_type: console format: html: code-fold: true code-tools: true --- [ Cetacean and sea turtle spatial density model outputs from visual observations using line-transect survey methods aboard NOAA vessel and aircraft platforms in the Gulf of Mexico from 2003-06-12 to 2019-07-31 (NCEI Accession 0256800) ](https://www.ncei.noaa.gov/access/metadata/landing-page/bin/iso?id=gov.noaa.nodc:256800) - additional [ metadata ](https://www.fisheries.noaa.gov/inport/item/67830) ```{r} #| messages: FALSE #| warning: FALSE # packages :: shelf (quiet = T)options (readr.show_col_types = F)source (here ("libs/db.R" )) # con <- function (con, tbl, flds, schema = "public" , geom= F, unique= F, overwrite= F, show= F, exec= T){# tbl = "taxa"; flds = c("tbl_orig", "aphia_id"); unique = T; geom=F stopifnot (! (geom == T & length (flds) != 1 ))<- ifelse (:: glue (" USING GIST ({flds})" ),:: glue ("({paste(flds, collapse=', ')})" ))<- ifelse (glue ("{tbl}_unique_idx" ),glue ("{tbl}_{paste(flds, collapse='_')}_idx" ))if (overwrite)dbExecute (con, glue ("DROP INDEX IF EXISTS {schema}.{idx}" ))<- glue:: glue ("CREATE {ifelse(unique, 'UNIQUE','')} INDEX IF NOT EXISTS {idx} ON {schema}.{tbl}{sfx}" )if (show)message (sql)if (exec)dbExecute (con, sql)``` ```{r d_datasets} <- "public" <- tibble (ds_key = "gm" ,name_short = "NOAA GoMex Cetacean & Sea Turtle Densities" , # short name in form of {source_broad} {regions} {taxa_groups} {response_type} name_full = "Cetacean and sea turtle spatial density model outputs from visual observations using line-transect survey methods aboard NOAA vessel and aircraft platforms in the Gulf of Mexico from 2003-06-12 to 2019-07-31 (NCEI Accession 0256800)" ,link = "https://www.ncei.noaa.gov/access/metadata/landing-page/bin/iso?id=gov.noaa.nodc:256800" ,citation = "Litz, Jenny; Aichinger Dias, Laura; Rappucci, Gina; Martinez, Anthony; Soldevilla, Melissa; Garrison, Lance; Mullin, Keith; Barry, Kevin; Foster, Marjorie (2022). Cetacean and sea turtle spatial density model outputs from visual observations using line-transect survey methods aboard NOAA vessel and aircraft platforms in the Gulf of Mexico from 2003-06-12 to 2019-07-31 (NCEI Accession 0256800). [indicate subset used]. NOAA National Centers for Environmental Information. Dataset. https://doi.org/10.25921/efv4-9z56. Accessed July 28, 2022." ,description = "Based on ship-based and aerial line-transect surveys conducted in the U.S. waters of the Gulf of Mexico between 2003 and 2019, the NOAA Southeast Fisheries Science Center (SEFSC) developed spatial density models (SDMs) for cetacean and sea turtle species for the entire Gulf of Mexico. SDMs were developed using a generalized additive modeling (GAM) framework to determine the relationship between species abundance and environmental variables (monthly averaged oceanographic conditions during 2015 - 2019). Models were extrapolated beyond the U.S. Gulf of Mexico to provide insight into potential high density areas throughout the Gulf of Mexico. However, extrapolations of this type should be interpreted with caution. This dataset includes 19 shapefiles for the SDMs for each cetacean and sea turtle species or species group." ,response_type = "density" , # response_type: one of: occurrence, range, suitability, probability or density taxa_groups = "cetacean, sea turtle" , # taxa_groups: one or more of: fish, invertebrates, marine mammals, cetaceans, sea turtles, seabirds, etc. year_pub = 2022 ,date_obs_beg = "2003-06-12" , date_obs_end = "2019-08-01" , # up to, but not including *_end date_env_beg = "2015-01-01" , date_env_end = "2020-01-01" , # up to, but not including *_end regions = list ("Gulf of Mexico" )) # array # names(d_dataset) |> paste(collapse = ", ") |> cat() <- Id (schema = schema, table = "sdm_datasets" )<- tbl (con, t_datasets) |> filter (ds_key == "gm" ) |> collect () |> nrow () == 1 if (! row_exists){# append, except array columns: regions dbWriteTable (append = T,|> select (- regions) )# append array column: regions <- glue ("ARRAY['{d_datasets$regions |> unlist() |> paste(collapse = ', ')}']" )dbExecute (glue ("UPDATE sdm_datasets SET regions = {regions_sql} WHERE ds_key = 'gm'" ) )tbl (con, t_datasets) |> filter (ds_key == "gm" ) |> collect () |> glimpse ()``` ```{r d_geoms} # helper functions ---- <- function (d_sf, i= 0 ){# i = 12; d_sf <- d$sf[[i]] message (glue ("{i} of {nrow(d_sf)} in get_annual_density() ~ {Sys.time()}" ))# get geometries <- d_sf %>% select (hexid = HEXID, geom = geometry)# get *_n abundance (#/40km2) per month and calculate annual average density (#/km2) <- glue ("{month.abb}_n" )<- d_sf %>% st_drop_geometry () %>% select (hexid = HEXID, any_of (mos_n)) %>% pivot_longer (- hexid, names_to = "mo_n" , values_to = "n" ) %>% filter (n != - 9999 ) %>% group_by (hexid) %>% summarize (n = mean (n),.groups = "drop" ) %>% mutate (density = n / 40 ) %>% select (hexid, density) # individuals / km2 # return geometries joined with summarized attributes <- geoms %>% left_join (by = "hexid" )# read layers ---- <- "/Users/bbest/My Drive/projects/offhab/data/raw/ncei.noaa.gov - GoMex cetacean & sea turtle SDMs/0256800/2.2/data/0-data/NOAA_SEFSC_Cetacean_SeaTurtle_SDM_shapefiles" <- glue ("{dir_shp}/../spp_gmx.xlsx" )<- read_excel (taxa_xls)<- Id (schema = schema, table = "sdm_geometries" )<- (tbl (con, t_geoms) |> filter (ds_key == "gm" ) |> collect () |> nrow () ) > 1 if (! rows_exist){<- tibble (path_shp = list.files (dir_shp, "shp$" , full.names= T)) |> mutate (base_shp = basename (path_shp) |> path_ext_remove (),taxa_shp = str_replace (base_shp, "(.*)_Monthly_.*" , " \\ 1" )) |> left_join (|> select (taxa_shp, taxa_sci, taxa_doc),by = "taxa_shp" ) |> mutate (population = map_chr (~ {if (str_detect (.x, "^(Oceanic)|(Shelf)" )) {return (str_replace (.x, "^(Oceanic|Shelf)_(.*)" , " \\ 1" ))else {return (NA )sf = map (path_shp, read_sf),d_density = imap (sf, get_annual_density),months_lst = map (sf, ~ {intersect (colnames (.),glue ("{month.abb}_n" )) %>% str_replace ("_n" , "" ) }),n_months = map_int (months_lst, length),months = map_chr (months_lst, paste, collapse= ";" ) )# get unique geometries <- d |> select (sf) |> mutate (sf = map (sf, \(x){ x |> select (gid = HEXID) } ) ) |> unnest (cols = c (sf)) |> distinct () |> arrange (gid) |> st_as_sf () |> # exclude "POLYGON" whole hexagon duplicates filter (st_geometry_type (geometry) == "MULTIPOLYGON" ) |> rename (geom_id = gid) |> mutate (geom_id = as.integer (geom_id),ds_key = "gm" ) |> relocate (ds_key, .before = geom_id) |> st_transform (4326 ) |> # from: USA_Contiguous_Lambert_Conformal_Conic # mapView(d_geoms) st_set_geometry ("geom" )st_write (d_geoms, con, t_geoms, append = T)<- "SELECT * FROM sdm_geometries WHERE ds_key = 'gm'" # st_read(con, query = q) |> # mapView() ``` ```{r d_vals} <- Id (schema = schema, table = "sdm_values" )<- tbl (con, t_vals) |> filter (ds_key == "gm" ) |> summarize (n ()) |> pull () > 0 if (! rows_exist){# get unique values <- d |> select (sp_key = taxa_sci, population, data = sf) |> mutate (data = map (data, \(x){|> st_drop_geometry () |> rename (geom_id = HEXID) |> pivot_longer (cols = - geom_id,names_to = "mo_var" ,values_to = "val" ) |> filter (val != - 9999 ) |> separate (col = mo_var,into = c ("mo" , "var" ),sep = "_" )|> unnest (cols = c (data)) |> mutate (ds_key = "gm" ) |> relocate (ds_key, .before = taxa_sci)# A tibble: 15,445,842 × 7 <- 2019 # using last year of obs/env so ISO 8601 compliant <- d_vals |> mutate (mm = setNames (1 : 12 , month.abb)[mo],mm_str = stringr:: str_pad (mm, 2 , pad = "0" ),# Time Interval notation using [ISO 8601])(https://en.wikipedia.org/wiki/ISO_8601) time_interval = glue ("{yr}-{mm_str}/P1M" )) |> select (- mo, - mm, - mm_str)# summarize d_vals to d_models <- d_vals |> group_by (|> summarize (.groups = "drop" ) |> rowid_to_column ("mdl_id" ) |> mutate (mdl_id = "gm" ) |> relocate (ds_key, .before = mdl_id)<- d_vals |> left_join (by = c ("ds_key" , "sp_key" , "population" , "time_interval" , "var" )) |> select (<- d_taxa |> group_by (taxa_sci) |> summarize (taxa_common = first (taxa_common) |> str_replace_all ("Oceanic " , "" ) ) |> mutate (ds_key = "gm" ) |> select (ds_key, sp_key = taxa_sci, sp_common = taxa_common) |> mutate (sp_scientific = sp_key)<- Id (schema = schema, table = "sdm_species" )<- tbl (con, t_spp) |> filter (ds_key == "gm" ) |> summarize (n ()) |> pull () > 0 if (! rows_exist)dbWriteTable (con, t_spp, d_spp, append = T)<- Id (schema = schema, table = "sdm_models" )<- tbl (con, t_mdls) |> filter (ds_key == "gm" ) |> summarize (n ()) |> pull () > 0 if (! rows_exist)dbWriteTable (con, t_mdls, d_mdls, append = T)# check: d_vals |> select(mo, time_interval) |> table() dbWriteTable (con, t_vals, d_vals, append = T)<- Id (schema = schema, table = "sdm_models" )tbl (con, t_mdls) |> select (- description) |> collect () |> datatable ()tbl (con, t_vals) |> head () |> collect () |> kable ()``` ```{r d_map} #| eval: FALSE # build query # tbl(con, t_mdls) |> # filter( # ds_key == "gm", # # sp_key == "Stenella frontalis", # # population == "Oceanic", # sp_key == "Balaenoptera ricei", # time_interval == "2019-01/P1M", # var == "n") |> # select(ds_key, mdl_id) |> # left_join( # tbl(con, t_vals), # by = c("ds_key", "mdl_id") ) |> # left_join( # tbl(con, t_geoms), # by = c("ds_key", "geom_id")) |> # select(val, geom) |> # show_query() # explain() <- " SELECT v.val, g.geom FROM ( SELECT ds_key, mdl_id FROM public.sdm_models WHERE ds_key = 'gm' AND -- sp_key = 'Stenella frontalis' AND -- population = 'Oceanic' AND sp_key = 'Balaenoptera ricei' AND time_interval = '2019-01/P1M' AND var = 'n' ) AS m LEFT JOIN public.sdm_values AS v ON ( m.ds_key = v.ds_key AND m.mdl_id = v.mdl_id ) LEFT JOIN public.sdm_geometries AS g ON ( m.ds_key = g.ds_key AND v.geom_id = g.geom_id )" <- st_read (con, query = q)mapView (p)``` 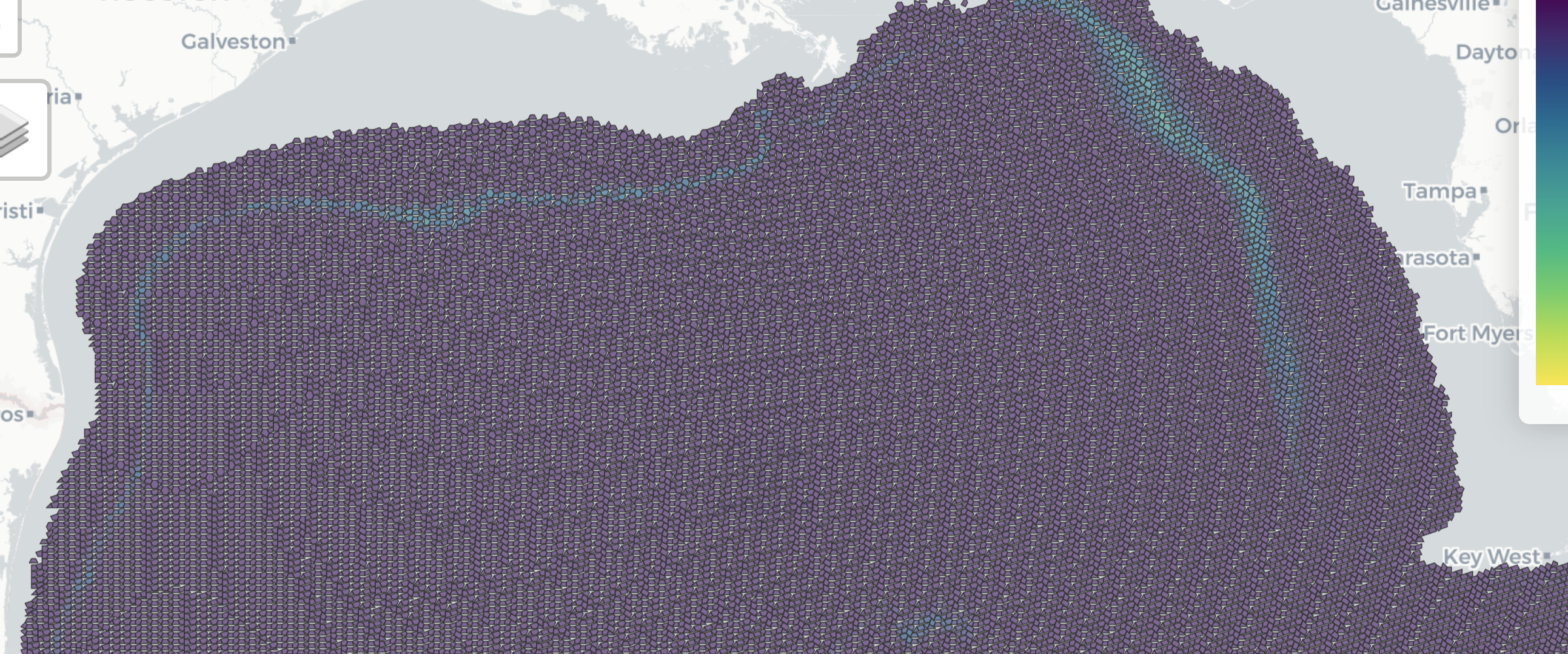 ```{r OLD} #| eval: FALSE <- d %>% select (mdl_id, taxa_sci, taxa_doc, months, n_months)<- d %>% select (%>% unnest (d_density)<- mdl_hex %>% group_by (hexid, geom) %>% summarize (.groups= "drop" ) %>% st_as_sf ()<- mdl_hex %>% st_drop_geometry () %>% select (mdl_id, hexid, density) %>% filter (! is.na (density))# helper functions ---- <- function (d_sf, i= 0 ){# i = 12; d_sf <- d$sf[[i]] message (glue ("{i} of {nrow(d)} in get_annual_density() ~ {Sys.time()}" ))# get geometries <- d_sf %>% select (hexid = HEXID, geom = geometry)# get *_n abundance (#/40km2) per month and calculate annual average density (#/km2) <- glue ("{month.abb}_n" )<- d_sf %>% st_drop_geometry () %>% select (hexid = HEXID, any_of (mos_n)) %>% pivot_longer (- hexid, names_to = "mo_n" , values_to = "n" ) %>% filter (n != - 9999 ) %>% group_by (hexid) %>% summarize (n = mean (n),.groups = "drop" ) %>% mutate (density = n / 40 ) %>% select (hexid, density) # individuals / km2 # return geometries joined with summarized attributes <- geoms %>% left_join (by = "hexid" )# add aphia_id ---- <- wm_add_aphia_id (gm_models, taxa_sci)# TODO ?: combine Oceanic_* & Shelf_* for AtlanticSpotted_Dolphin + CommonBottlenose_Dolphin # write to database ---- <- oh_pg_con ()dbWriteTable (con, "gm_models" , gm_models, overwrite= T)st_write (gm_model_hexagons, con, "gm_model_hexagons" , delete_layer= T)dbWriteTable (con, "gm_model_hexagon_densities" , gm_model_hexagon_densities, overwrite= T)create_index (con, "gm_models" , "mdl_id" , unique = T)create_index (con, "gm_model_hexagons" , "hexid" , unique = T)create_index (con, "gm_model_hexagon_densities" , c ("mdl_id" , "hexid" ), unique = T)# hexid x cell_id ---- tbl (con, "gm_model_hexagons" )<- dbGetQuery ("WITH c AS ( SELECT cell_id, geom FROM oh_cells), d AS ( SELECT hexid AS ply_id, ST_Transform(geom, 4326) AS geom FROM gm_model_hexagons) SELECT DISTINCT ON (tbl, ply_id, cell_id) 'gm_model_hexagons' AS tbl, d.ply_id, c.cell_id FROM c JOIN d ON ST_Covers(d.geom, c.geom)" ) %>% tibble ()dbSendQuery (con, "DELETE FROM oh_cells_ply WHERE tbl = 'gm_model_hexagons'" )dbAppendTable (con, "oh_cells_ply" , oh_cells_ply)# test model output ---- <- oh_pg_con ()<- tbl (con, "gm_models" ) %>% filter (taxa_sci == "Ziphius" ) %>% left_join (tbl (con, "gm_model_hexagon_densities" ),by = "mdl_id" ) %>% left_join (tbl (con, "oh_cells_ply" ) %>% filter (tbl == "gm_model_hexagons" ),by = c ("hexid" = "ply_id" )) %>% select (cell_id, density) %>% filter ( # 1,977,010 -> 1,959,979 ! is.na (cell_id),! is.na (density)) %>% collect ()library (terra)<- oh_rast ()$ cell_id] <- r_d$ density<- tempfile (fileext = ".tif" )<- trim (r)plot (r)mapView (r, maxpixels= ncell (r))# redo rast ---- :: shelf (options (readr.show_col_types = F)<- "/Users/bbest/My Drive/projects/offhab/data/derived/lyrs_tif" <- here ("data-raw/layers.csv" )<- "gm" # match with aphia_id <- d |> left_join (tbl (con, "taxa" ) |> # distinct(tbl) |> pull(tbl) filter (tbl == "gm_models" ) |> select (taxa_sci = taxa,|> collect (),by = "taxa_sci" )# combine: # Stenella frontalis | Tursiops truncatus # oceanic | 8 | 9 # shelf | 15 | 16 <- D |> filter (== 8 ) |> select (aphia_id, d_density) |> unnest (d_density) |> rename (density_oceanic = density) |> left_join (|> filter (== 15 ) |> select (mdl_id, d_density) |> unnest (d_density) |> select (density_shelf = density),by = "hexid" ) |> mutate (density = map2_dbl (na.rm = T)) |> select (- density_oceanic, - density_shelf) |> nest (d_density = c (hexid, geom, density))<- D |> filter (== 9 ) |> select (aphia_id, d_density) |> unnest (d_density) |> rename (density_oceanic = density) |> left_join (|> filter (== 16 ) |> select (mdl_id, d_density) |> unnest (d_density) |> select (density_shelf = density),by = "hexid" ) |> mutate (density = map2_dbl (na.rm = T)) |> select (- density_oceanic, - density_shelf) |> nest (d_density = c (hexid, geom, density))<- D |> filter (! mdl_id %in% c (8 ,9 ,15 ,16 )) |> select (aphia_id, d_density) |> rbind (# iterate over rasters <- oh_rast ("NA" )# for (i in 1:nrow(D)){ # i = 1 for (i in 16 : nrow (D)){ # i = 16 <- D$ aphia_id[i]<- glue ("{ds_key}_{aphia_id}" )<- glue ("{dir_lyrs_tif}/{lyr_key}.tif" )message (glue ("{i} of {nrow(D)}: {basename(r_tif)} ~ {Sys.time()}" ))# get density, geom <- D |> slice (i) |> pull (d_density) %>% 1 ]] |> st_as_sf ()# mapView(d, zcol = "density") # convert to raster <- d |> st_transform (3857 ) |> rasterize ("density" )# plet(r, tiles="Esri.NatGeoWorldMap") # rescale 0 to 100; set 0 -> NA <- range (values (r, na.rm = T)))<- setValues (:: rescale (values (r),to = c (0 , 100 )) )== 0 ] <- NA # plet(r, tiles="Esri.NatGeoWorldMap") # write raster write_rast (r, r_tif)# write min, max to layers.csv <- read_csv (lyrs_csv) |> filter (!= !! lyr_key) |> bind_rows (tibble (lyr_key = lyr_key,ds_key = ds_key,aphia_id = aphia_id,val_min = vr[1 ],val_max = vr[2 ],rescale_min = 0 ,rescale_max = 100 ) )# message(glue(" nrow(lyrs_csv): {nrow(d)}")) write_csv (d, lyrs_csv)``` 