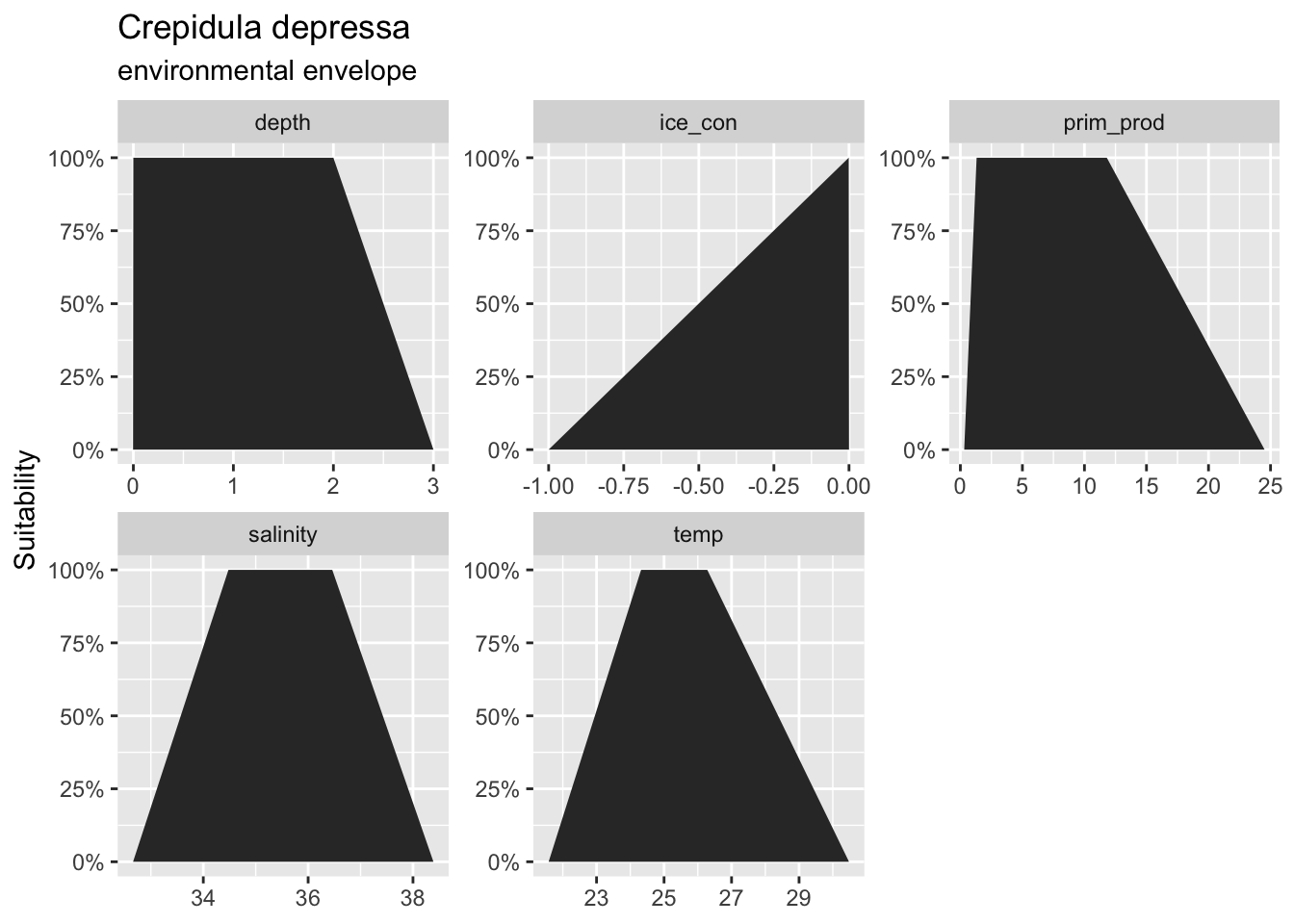
--- title: "Explore Interpolation of SDMs" editor: visual execute: warning: false editor_options: chunk_output_type: console --- - AquaMaps raster from env REsuitability - Get a few species with different geographic traits, starting with species in Gulf of America: - small range, high density - large range, low density- Bottom Trawl from IDW- Other: GoMex hexagons from complex SDM- raster: - 1/12° (0.083°)\ - 1/20° (0.050°)\- hexagon: - Uber H3- "chunky": interpolate from underlying values- "smooth": interpolate from centroid points- "remap": re-ramp environmental envelope from higher resolution data - only AquaMaps, possibly DisMAP- [ MarineSDMs - Explorations ](https://marinebon.github.io/MarineSDMs/explorations.html) - [ MarineSDMs - AquaMaps Downscaled ](https://marinebon.github.io/MarineSDMs/explorations/am-fine.html) - [ marinebon/aquamaps-downscaled: initial attempt to downscale AquaMaps species distribution models to finer spatial resolution a la GEBCO bathymetry (15 arc second) ](https://github.com/marinebon/aquamaps-downscaled/tree/main) - [ aquamaps-downscaled/index.qmd at main · marinebon/aquamaps-downscaled ](https://github.com/marinebon/aquamaps-downscaled/blob/503c62f39552c4ad2417413b13c49d4d4405f3ab/index.qmd) - [ Downscaling AquaMaps ](https://marinebon.github.io/aquamaps-downscaled/) - [ aquamaps-downscaled/am-env\_to\_ee.qmd at main · marinebon/aquamaps-downscaled ](https://github.com/marinebon/aquamaps-downscaled/blob/main/am-env_to_ee.qmd) ## AquaMaps duckdb database ```{r} #| label: setup :: shelf (# raquamaps/raquamaps, / msens,quiet = T) # zeallot mapviewOptions (basemaps = "Esri.OceanBasemap" )<- "~/My Drive/projects/msens/data" <- glue ("{dir_data}/derived/aquamaps/am.duckdb" )<- dbConnect (duckdb (dbdir = path_dd,read_only = T))# dbDisconnect(con_dd, shutdown = T) # dbListTables(con_dd) # "_tbl_fld_renames" "cells" "spp" "spp_cells" "spp_occs" "spp_prefs" tbl (con_dd, "_tbl_fld_renames" ) |> collect () |> datatable ()``` ## Gulf of America [ `msens` ](https://marinesensitivity.org/msens/news/index.html#msens-020) : `ply_boemrgns` \> `ply_ecorgns` \| `ply_planareas` \> `ply_ecoareas` - [ ] Rename Gulf of Mexico to Gulf of America - `boemrgn_key` : "brGOM" -\> "brGOA" - `boemrgn_name` : "Gulf of Mexico" -\> "Gulf of America"```{r} #| label: spp_goa # st_bbox(msens::ply_boemrgns) # xmin ymin xmax ymax # 166.95776 23.78077 295.89167 74.99636 # so [0,360], not [-180, 180] <- msens:: ply_boemrgns |> filter (boemrgn_key == "brGOM" ) |> st_wrap_dateline () # [ 0, 360] -> [-180, 180] # st_shift_longitude() # [-180, 180] -> [ 0,360] # st_bbox(goa) # xmin ymin xmax ymax # -97.23899 23.78077 -81.17011 30.28910 mapView (goa)``` ## Get species in Gulf of America ```{r} # get cells within goa <- tbl (con_dd, "cells" ) |> collect () |> st_as_sf (coords = c ("center_long" , "center_lat" ), crs = 4326 , remove = F)<- st_intersection (pt_cell, goa)# get species in gom <- tbl (con_dd, "spp_cells" ) |> filter (cell_id %in% pt_cell_goa$ cell_id) |> group_by (sp_key) |> collect ()# get species ranges by n_cells <- tbl (con_dd, "spp_cells" ) |> filter (sp_key %in% d_sp_goa$ sp_key) |> group_by (sp_key) |> summarize (n_cells = n (), .groups = "drop" ) |> left_join (tbl (con_dd, "spp" ), by = "sp_key" ) |> arrange (n_cells) |> collect ()bind_rows (slice (d_sp_goa_rng, 1 : 100 ),slice (d_sp_goa_rng, (n () - 99 ): n ())) |> datatable (caption = htmltools:: tags$ caption (style = 'caption-side: top; text-align: left;' ,withTags (div (HTML (glue (" Table. <i>Species in Gulf of America (n = {format(nrow(d_sp_goa_rng), big.mark=',')}), subset to first 100 species and last 100 species, sorted by range size (n_cells).</i>" ))))))``` ## Get AquaMaps env data ```{r} #| label: am_hcaf_env # source: https://github.com/marinebon/aquamaps-downscaled/blob/main/am-env_to_ee.qmd <- here ("data/am-hcaf.tif" )if (! file.exists (h_tif)){# get aquamaps table of env values ---- # d_cells <- tbl(con_dd, "cells") |> # collect() # raster template ---- # range(d$ctr_lon) # -179.75 179.75 # range(d$ctr_lat) # - 78.25 89.75 # r_template <- rast( # xmin = min(d_cells$center_long) - 0.25, # xmax = max(d_cells$center_long) + 0.25, # ymin = min(d_cells$center_lat) - 0.25, # ymax = max(d_cells$center_lat) + 0.25, # resolution = c(0.5, 0.5), # crs = "epsg:4326") # Create the global template raster <- - 180 ; xmax <- 180 <- - 90 ; ymax <- 90 <- 0.5 # Half-degree resolution <- rast (nrows = (ymax - ymin) / res,ncols = (xmax - xmin) / res,xmin = xmin,xmax = xmax,ymin = ymin,ymax = ymax,crs = "EPSG:4326" )# data frame to points ---- <- tbl (con_dd, "cells" ) |> collect () |> st_as_sf (coords = c ("center_long" , "center_lat" ),remove = F,crs = 4326 ) |> arrange (center_long, center_lat)# points to raster ---- <- function (v){<- rasterize (p, r_g, field = v)names (r) <- v<- NULL for (v in setdiff (names (p), "geometry" )){message (glue ("v: {v}" ))if (is.null (r_h)){<- get_r (v)else {<- rast (list (r_h, get_r (v)))writeRaster (r_h, h_tif, overwrite= T)<- rast (h_tif)# names(r_h) # [1] "cell_id" "cell_idx" "csquare_code" "loiczid" # [5] "n_limit" "s_limit" "w_limit" "e_limit" # [9] "center_lat" "center_long" "cell_area" "ocean_area" # [13] "p_water" "clim_zone_code" "fao_area_m" "fao_area_in" # [17] "country_main" "country_second" "country_third" "country_sub_main" # [21] "country_sub_second" "country_sub_third" "eez" "lme" # [25] "lme_border" "meow" "ocean_basin" "islands_no" # [29] "area0_20" "area20_40" "area40_60" "area60_80" # [33] "area80_100" "area_below100" "elevation_min" "elevation_max" # [37] "elevation_mean" "elevation_sd" "depth_min" "depth_max" # [41] "depth_mean" "depth_sd" "sst_an_mean" "sbt_an_mean" # [45] "salinity_mean" "salinity_b_mean" "prim_prod_mean" "ice_con_ann" # [49] "oxy_mean" "oxy_b_mean" "land_dist" "shelf" # [53] "slope" "abyssal" "tidal_range" "coral" # [57] "estuary" "seamount" "mpa" # plot to check # static: # plot(r_h[["center_lat"]]) # plot(r_h[["center_long"]]) # dynamic: # plet(r_h[["center_lat"]]) # plet(r_h[["center_long"]]) <- "depth_mean" ; clr <- "dense" mapView (layer.name = lyr,col.regions = cmocean (clr),alpha.regions = 0.9 )<- "prim_prod_mean" ; clr <- "algae" mapView (layer.name = lyr,col.regions = cmocean (clr),alpha.regions = 0.9 )<- "sst_an_mean" ; clr <- "thermal" mapView (layer.name = lyr,col.regions = cmocean (clr),alpha.regions = 0.9 )<- "sbt_an_mean" ; clr <- "thermal" mapView (layer.name = lyr,col.regions = cmocean (clr),alpha.regions = 0.9 )``` ## Get species attributes - [ Computer Generated Native Distribution Map for _Crepidula depressa_ ](https://aquamaps.org/preMap2.php?cache=1&SpecID=W-Msc-419703) 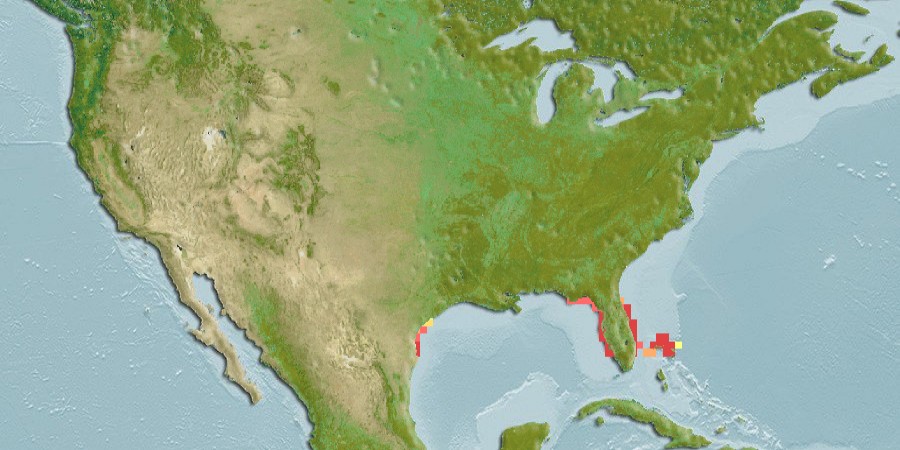 - [ Mapping Parameters ](https://www.aquamaps.org/ShowMapParam.php?SpecID=W-Msc-419703&graph=&user_session=1&bp=-1) - `"W-Msc-419703"` : _Crepidula depressa_ (class = Gastropoda) n_cells = 50```{r} #| label: get_sp_pref <- "W-Msc-419703" <- tbl (con_dd, "spp_prefs" ) |> filter (sp_key == !! sp_key) |> collect ()|> mutate (across (everything (), as.character)) |> pivot_longer (everything ()) |> kable ()``` - [ ] Reconcile differences in bbox and FAO areas vs online.```{r} :: shelf (<- function (sp_key){# source: https://github.com/marinebon/aquamaps-downscaled/blob/503c62f39552c4ad2417413b13c49d4d4405f3ab/sp-map/functions.R#L92C5-L93C30 <- tbl (con_dd, "spp_prefs" ) |> filter (sp_key == !! sp_key) |> collect ()<- d_prefs |> select (ends_with ("_yn" )) |> pivot_longer (everything ()) |> filter (value == 1 ) |> pull (name) |> str_replace ("_yn$" ,"" ) |> setdiff ("extn_rule" )<- tribble (~ prob_name, ~ prob_value,"min" , 0 ,"pref_min" , 1 ,"pref_max" , 1 ,"max" , 0 )# browser() <- d_prefs |> select (starts_with (vars_yes)) |> select (! ends_with ("_yn" )) |> pivot_longer (everything (),values_to = "var_value" ) |> separate_wider_regex (c (var = paste (vars_yes, collapse = "|" ),"_" ,prob_name = paste (d_probs$ prob_name, collapse = "|" ))) |> left_join (by = "prob_name" )<- d_env |> group_by (var) |> summarise (vec = list (var_value)) |> deframe ()<- list ()<- tbl (con_dd, "spp" ) |> filter (sp_key == !! sp_key) |> collect () |> mutate (sp_sci = glue ("{genus} {species}" ))"sp_scientific" ] <- list (d_sp$ sp_sci)"sp_key" ] <- list (d_sp$ sp_key)"sp_int" ] <- list (d_sp$ sp_int)"fao_areas" ] <- d_prefs$ fao_areas |> str_split (", \\ s*" ) |> str_trim () |> unlist () |> as.numeric () |> sort ()"env" ] <- list (l_env)$ taxa <- d_sp |> select (kingdom, phylum, class, order, family) |> pivot_longer (everything ()) |> deframe () |> as.list ()$ attr <- d_sp |> select (|> mutate (across (everything (), as.character)) |> pivot_longer (everything ()) |> deframe () |> as.list ()$ iucn <- d_sp |> select (|> mutate (across (everything (), as.character)) |> pivot_longer (everything ()) |> deframe () |> as.list ()# deepwater, # Does the species occur in the deep-sea (i.e. tagged bathypelagic or bathydemersal in FishBase or SeaLifeBase)? 0=No, 1=Yes # angling, # Is the species a sport fish (i.e. tagged as a GameFish in FishBase)? 0=No, 1=Yes # diving, # Is the species found on a dive (i.e. where DepthPrefMin in HSPEN < 20 meters)? 0=No, 1=Yes # dangerous, # Is the species dangerous (i.e. tagged as ‘traumatogenic or venonous’ in FishBase or SeaLifeBase)? 0=No, 1=Yes # m_invertebrates, # Is the species a marine invertebrate? 0=No, 1=Yes # highseas, # Is the species an open ocean fish species (i.e. tagged as pelagic-oceanic in FishBase)? 0=No, 1=Yes # invasive, # Is the species recorded to be invasive (i.e. in FishBase or SeaLifeBase)? 0=No, 1=Yes # resilience, # Resilience of the species (i.e. as recorded in FishBase/SeaLifeBase) [varchar] # iucn_id, # IUCN species identifier [int] # iucn_code, # IUCN Red list classification assigned to the species [varchar] # iucn_version, # IUCN version [varchar] # provider) |> # FishBase (FB) or SeaLifeBase (SLB)? [varchar] <- get_sp_info (sp_key)<- function (sp_info){<- sp_info$ env |> enframe (name = "variable" ) |> mutate (suitability = list (c (0 ,1 ,1 ,0 ))) |> unnest (c (value, suitability))<- ggplot (d, aes (value, suitability)) + geom_area () + scale_y_continuous (labels = percent) + facet_wrap (vars (variable), scales = "free" ) + labs (title = sp_info$ sp_scientific,subtitle = "environmental envelope" ,x = NULL ,y = "Suitability" ) + theme_gray ()plot_sp_env (sp_info)jsonedit (mode = "view" , modes = c ("view" ,"code" ))``` ## Compare with native `aquamapsdata` ```{r} # dependency for aquamapsdata: # - Terminal: brew install gnupg :: shelf (/ rcrypt,/ aquamapsdata,# initial run-once step required to install remote db locally # download_db(force = TRUE) default_db ("sqlite" )# get the identifier for the species <- am_raster (sp_key)<- ext (ras) |> as.vector () |> as.numeric ()# show the native habitat map am_map_leaflet (ras, title = sp_key) |> :: fitBounds (lng1 = b[1 ], lat1 = b[3 ], lng2 = b[2 ], lat2 = b[4 ])# xmin xmax ymin ymax # -97.75 -76.75 26.25 29.75 # use one or more keys for species am_hspen () |> filter (SpeciesID == sp_key) |> head (1 ) |> collapse () |> glimpse ()# $ FAOAreas <chr> "31" # $ FAOComplete <int> NA # $ NMostLat <dbl> 30 # $ SMostLat <dbl> 26 # $ WMostLong <dbl> 97 # $ EMostLong <dbl> 80 ``` ## Custom Leaflet functions ```{r} #| label: custom leaflet functions <- function (m){# m: leaflet() map |> # add base: blue bathymetry and light brown/green topography addProviderTiles ("Esri.OceanBasemap" ,options = providerTileOptions (variant = "Ocean/World_Ocean_Base" )) |> # add reference: placename labels and borders addProviderTiles ("Esri.OceanBasemap" ,options = providerTileOptions (variant = "Ocean/World_Ocean_Reference" ))# show the native habitat map <- function (id = "am_rast" ,cols = c ("#FEB24C" , "#FD8D3C" , "#FC4E2A" , "#E31A1C" , "#B10026" ),truncate_to_zero = T){# m: leaflet() map # r: raster # TODO : migrate to terra::rast() # r = r_gebco_bb # title = "GEBCO depth (m)" # cols = RColorBrewer::brewer.pal(7, "Blues") <- leaflet:: projectRasterForLeaflet (r, method = "bilinear" )# truncate to 0 to prevent negative values # that were generated by projecting the raster # from geographic projection (decimal degrees) to Web Mercator (meters) if (truncate_to_zero){<- values (r)< 0 ] <- 0 values (r) <- v<- leaflet:: colorBin (na.omit (unique (values (r))), bins = length (cols), pretty = TRUE , na.color = "#00000000" )<- terra:: ext (r) |> :: st_bbox () |> st_as_sfc () |> st_as_sf (crs= 3857 ) |> st_transform (4326 ) |> st_bbox ()|> :: addRasterImage (layerId = id, group = id,project = F, colors = pal, opacity = 0.8 ) |> :: addLegend (values = raster:: values (r), title = title, pal = pal) |> :: fitBounds (lng1 = e[["xmin" ]], lat1 = e[["ymin" ]], lng2 = e[["xmax" ]], lat2 = e[["ymax" ]]) |> addImageQuery (layerId = id, project = F) |> addLayersControl (overlayGroups = id)``` ```{r} :: shelf (<- function (lbl_left = "native →" , lbl_right = "← replicated" ,legend_title = "AquaMaps<br>suitability" ){= RColorBrewer:: brewer.pal (5 , "YlOrRd" )<- leaflet:: colorBin (c (0 , 1 ), bins = length (cols), pretty = TRUE , na.color = "transparent" )leaflet (options = leafletOptions (attributionControl = F,zoomControl = F)) |> addMapPane ("left" , zIndex = 1 ) |> addMapPane ("right" , zIndex = 1 ) |> addProviderTiles ($ Esri.OceanBasemap,group = "base" , layerId = "base_left" , options = pathOptions (pane = "left" )) |> addProviderTiles ($ Esri.OceanBasemap,group = "base" , layerId = "base_right" , options = pathOptions (pane = "right" )) |> addRasterImage (colors = pal, opacity = 0.8 , options = leafletOptions (pane = "left" ), group = lbl_left) |> addRasterImage (colors = pal, opacity = 0.8 , options = leafletOptions (pane = "right" ), group = lbl_right) |> addLegend (values = seq (0 , 1 , length.out = 10 ), pal = pal,title = legend_title,position = "bottomright" ,labFormat = \(x, type){<- x[1 : length (x)- 1 ]* 100 <- x[2 : length (x)]* 100 glue ("{str_pad(b,2)} - {e}%" )}) |> addControl (lbl_left, position = "topleft" ) |> addControl (lbl_right, position = "topright" ) |> # addLayersControl( # overlayGroups = c(lbl_left, lbl_right)) |> addSidebyside (layerId = "sidecontrols" ,rightId = "base_right" ,leftId = "base_left" )``` [ Content - Introduction to aquamapsdata • aquamapsdata ](https://raquamaps.github.io/aquamapsdata/articles/intro.html#content) - `depth/temp/salinity/prim_prod/ice_con/oxy/land_dist_yn` : Is the depth/temperature/salinity primary production/ice concentration/oxygen/distance to land parameter used in computing map data? 0=No, 1=Yes- `layer` : Indicates whether the **temperature** and **salinity** parameters are based on ***bottom*** (=b) or ***surface*** (=s) values of half-degree cells used to compute the envelope.- `spp_prefs.map_opt` : Indicates how native map (predicted probabilities) is plotted: 1 = area covered by both species’ bounding box and FAO areas, 2 = area covered by species’ FAO areas only, 3 = area covered by species’ bounding box only. - `spp_prefs.extn_rule_yn` : Was the FAO extension rule applied in the generation of the species envelope? 0=No, 1=Yes, null - `spp_cells.fao_area_yn` : Does this cell fall within an FAO area where the species is known to occur (endemic/native)? 0=No, 1=Yes - `spp_cells.bound_box_yn` : Does this cell fall within the geographical bounding box known for the species? 0=No, 1=Yes- `mean_depth` : Is mean depth used to fit the depth envelope? By default, marine mammals use mean depth. 0=No, 1=Yes- [ ] inspect all "pelagic" references - `pelagic` : Does the species occurs in the water column well above and largely independent of the bottom? 0=No, 1=Yes - `deepwater` : Does the species occur in the deep-sea (i.e. tagged bathypelagic or bathydemersal in FishBase or SeaLifeBase)? 0=No, 1=Yes - `highseas` : Is the species an open ocean fish species (i.e. tagged as pelagic-oceanic in FishBase)? 0=No, 1=Yes- `diving` : Is the species found on a dive (i.e. where DepthPrefMin in HSPEN < 20 meters)? 0=No, 1=Yes ## Get species raster ```{r} <- tbl (con_dd, "spp_cells" ) |> filter (sp_key == !! sp_key) |> collect ()# d_sp_cell # # A tibble: 50 × 5 # sp_key probability fao_area_yn bound_box_yn cell_id # <chr> <dbl> <int> <int> <int> # 1 W-Msc-419703 0.18 1 1 211899 # 2 W-Msc-419703 0.96 1 1 211900 # 3 W-Msc-419703 0.85 1 1 211901 # 4 W-Msc-419703 1 1 1 211902 # table(d_sp_cell$fao_area_yn, d_sp_cell$bound_box_yn, useNA = "always") # 1 <NA> # 1 50 0 # <NA> 0 0 <- tbl (con_dd, "cells" ) |> filter (cell_id %in% d_sp_cell$ cell_id) |> collect ()# View(d) # range(d_sp_cell$probability) # 0.03 1.00 # c(min(d$w_limit), max(d$e_limit), min(d$s_limit), max(d$n_limit)) # -98.0 -76.5 26.0 30.0 # get_sp(sp_key) ---- <- with (subst ( # substitute cell_id with probability "cell_id" ]], from = cell_id,to = probability,others = NA )) |> trim ()names (r_sp) <- "probability" # ext(r_sp) # -98, -76.5, 26, 30 (xmin, xmax, ymin, ymax) <- "YlOrRd" <- colorRampPalette (brewer.pal (n= 5 , name= pal_col))<- c (0 , 0.2 , 0.4 , 0.6 , 0.8 , 1 )<- 0.9 plet (r_sp, col = pal_col, tiles = "Esri.OceanBasemap" )``` ## Get species raster with env as diagnostic polygons ```{r} <- c (|> crop (r_sp) |> mask (r_sp))<- terra:: as.polygons ("cell_id" ]]) |> st_as_sf () |> left_join (as.data.frame (r_sp_h),by = "cell_id" ) |> relocate (probability) |> as_tibble () |> st_as_sf ()# apply yellow to red color ramp to probability mapView (zcol = "probability" , col.regions = pal_rmp, at = pal_at, alpha.regions = pal_alpha)``` - [ ] Limit ply attrs to relevant fields, esp env values and RES min/max## Develop raster topology - 1/12° (`r round(1/12, 4)` °: `r 360 * 1/12` x `r 90 * 1/12` ) -- Copernicus Marine- 1/20° (`r round(1/20, 4)` °: `r 360 * 1/20` x `r 90 * 1/20` ) -- Bio-Oracle (new sdmpredictors)```{r} #| label: topo_raster <- 1 / 12 <- rast (xmin = - 180 , xmax = 180 , ymin = - 90 , ymax = 90 , resolution = res, crs = "EPSG:4326" )``` ## Resample nearest `terra::resample(method = "near")` to get the higher resolution data using nearest neighbor.```{r} <- resample (method = "near" ) |> trim ()names (r_sp_n) <- "pr_near" plet (r_sp_n, col = pal_col, tiles = "Esri.OceanBasemap" )``` ## Resample bilinear `terra::resample(method = "bilinear")` to get the higher resolution data using bilinear interpolation.```{r} <- resample (method = "bilinear" ) |> trim ()names (r_sp_b) <- "pr_bilinear" plet (r_sp_b, col = pal_col, tiles = "Esri.OceanBasemap" )``` ## Reinterpolate - ```{r} <- as.points (r_sp) |> st_as_sf ()# mapView(pts_sp, cex = 2) <- as.polygons (r_sp > 0 ) |> st_as_sf ()# mapView(ply_sp, cex = 2) <- res (r_sp)[1 ]<- setValues (r_sp |> extend (1 ), 0 )sf_use_s2 (F)<- ply_sp |> st_buffer (w) |> st_union () |> st_make_valid () |> st_as_sf () |> st_difference (ply_sp)sf_use_s2 (T)# mapView(ply_0, cex = 2) <- r_0 |> crop (ply_0) |> mask (ply_0) |> as.points () |> st_as_sf ()names (pts_0)[1 ] <- names (pts_sp)[1 ]# mapView(pts_sp, cex = 4) + # mapView(pts_0, cex = 2, col.regions = "gray") <- bind_rows (= 2 = w # to include cardinal neighbors, w * sqrt(2) ~= w * 1.4 = 0.1 # 10% min <- interpIDW (x = r_g |> crop (r_0), y = pts_sp0 |> vect (), field = names (pts_sp0)[1 ],radius = rad, # 2/3, power = pwr) # /2) <- mask (r_idw, r_idw >= val_min, maskvalue = F) |> trim ()# plot( # r_idw, # main = glue("idw(radius = {rad}, power = {pwr})")) <- mapView (r_idw) + mapView (pts_sp, cex = 4 ) + mapView (pts_0, cex = 2 , col.regions = "gray" )@ map |> addFullscreenControl ()``` ## Mask by land ```{r} :: shelf (quiet = T)# if (!requireNamespace("rnaturalearthhires", quietly = TRUE)) { # install.packages( # "rnaturalearthhires", # repos = "https://ropensci.r-universe.dev", # type = "source") # } # library(rnaturalearthhires) # ne_states_x <- rnaturalearthhires::states10 |> <- rnaturalearthdata:: states50 |> # ne_states_x <- rnaturalearth::ne_states() |> st_make_valid () |> st_filter (ext (r_idw) |> vect () |> st_as_sf () |> st_set_crs (4326 ),.predicate = st_intersects) |> tibble () |> st_as_sf ()# ne_states_x_0 <- ne_states_x # ne_states_x <- ne_states_x |> # st_transform(st_crs(r_idw)) |> # tibble() |> # st_as_sf() = 0.95 <- rasterize (|> mutate (one = 1 ),"one" , cover = T)# r_land <- mask( # r_land, # r_land > pct_land, # maskvalue = F) # plot(r_land) #, main = glue("land (pct = {pct_land})")) # plot(r_land >= pct_land) #, main = glue("land (pct = {pct_land})")) <- mask (> pct_land, maskvalue = T)# plot(r_idw_m) # plot(r_idw) # mapView(r_idw) + mapView (r_idw_m) + mapView (ne_states_x, col.regions = "gray" , alpha = 0 )``` ## Replicate AquaMaps species Probability from env layers ```{r} # parameters # sp_key: # TODO : fetch d_sp_pref based on sp_key <- d_sp_pref |> select (ends_with ("_yn" )) |> # names() |> # |> pivot_longer (everything ()) |> filter (value == 1 ) |> pull (name) |> str_replace ("_yn" , "" )# "depth" "temp" "salinity" "prim_prod" "ice_con" "oxy" "land_dist" "extn_rule" <- r_h# bbox ---- <- d_sp_pref$ map_opt %in% c (1 ,3 )if (bbox_yes){# OLD bbox: off # bbox_ext <- d_sp_pref |> # # 45 -28 -95 -33 # select(w_most_long, e_most_long, s_most_lat, n_most_lat) |> # mutate( # w_most_long = w_most_long * -1, # e_most_long = e_most_long * -1) |> # pivot_longer(everything()) |> # deframe() |> # as.vector() |> # ext() # NEW bbox: from species occurrences # goal: approximate native r_sp_old vs r_sp_new; using spp_occs # r_sp_old: -98 , -76.5 , 26 , 30 (xmin, xmax, ymin, ymax) # r_sp_new: -97 , -80 , 26 , 30 # pts_occs: -97.25, -77.25, 23.25, 30.25 # pts_good: -97.25, -77.25, 26.25, 29.25 # pts_ibnd: -97.25, -77.25, 26.25, 29.25 # bb_occs: -97.5 , -77.0 , 26.0 , 29.5 # bb_pad: -98.0, -76.5 , 25.5 , 30.0 <- 0.5 # bb_pad <- tbl (con_dd, "spp_occs" ) |> filter (== !! sp_key,== 1 ) |> left_join (tbl (con_dd, "cells" ),by = "cell_id" ) |> summarize (xmin = min (w_limit, na.rm = T) - pad,xmax = max (e_limit, na.rm = T) + pad,# ymin = min(s_limit, na.rm = T) - pad, ymin = min (s_limit, na.rm = T),ymax = max (n_limit, na.rm = T) + pad) |> collect () |> pivot_longer (everything ()) |> deframe () |> as.vector () |> ext ()<- crop (r_env, bbox_ext)# fao ---- if (("extn_rule" %in% vars_yes | $ map_opt %in% c (1 ,2 )) && ! is.na (d_sp_pref$ fao_areas)){# d_fao <- tbl(con_dd, "spp_prefs") |> # distinct(fao_areas) |> # collect() |> # mutate( # n = str_split(fao_areas, ",\\s*") |> # purrr::map_dbl(length)) |> # arrange(n) # View(d_fao) # Conclusion: all species have at least 1 FAO area # example: fao_areas = "57, 61, 71" <- d_sp_pref$ fao_areas |> str_split (", \\ s*" ) |> str_trim () |> unlist () |> as.numeric () |> sort ()<- r_env[["fao_area_m" ]] %in% fao_areas<- mask (maskvalue = F)<- setdiff (vars_yes, "extn_rule" )<- function (v, p){# p = c(-1, 0, 0, 0) # dput(p) for var = "ice_con"; sp_key = "W-Msc-419703" # v = 0 approx (x = p, # c(min, min_pref, max, max_pref) y = c (0 , 1 , 1 , 0 ), xout = v, # yleft = 0, # v < y_min -> 0 # yright = 0, # v > y_max -> 0 # rule = 2, # 2: return nearest vs 1: NA rule = 1 , # return NA if outside range method = "linear" ,ties = max)$ y# ties = "ordered")$y # assume x is ordered # match species preference var to AquaMaps env raster layers <- d_sp_pref$ layer == "s" # vs "b" bottom <- list ("depth" = "depth_mean" ,# TODO : use depth_min, depth_max, depth_sd? "temp" = ifelse ("sst_an_mean" ,"sbt_an_mean" ),"salinity" = ifelse ("salinity_mean" ,"salinity_b_mean" ),# "oxy" = ifelse( # is_surface, # "oxy_mean", # "oxy_b_mean"), "oxy" = "oxy_b_mean" ,"prim_prod" = "prim_prod_mean" ,"ice_con" = "ice_con_ann" ,"land_dist" = "land_dist" )<- list ()for (var in vars_yes){ # var = vars_yes[1] # var = "depth" # var = "prim_prod" # get Relative Env Suitability (res) parameters <- d_sp_pref |> select (sprintf ("%s_%s" , var, c ("min" , "pref_min" , "pref_max" , "max" ))) |> pivot_longer (everything ()) |> deframe () |> as.vector ()# check that p is ordered stopifnot (length (p) == 4 ,all (is.finite (p)),all (diff (p) >= 0 ))<- var_lyr[[var]]<- r_env[[lyr]]# plot(r_var) <- terra:: app (x = r_var, fun = ramp_env, p = p)# plot(r_sp_env[[var]]) # mapView(r_sp_env[[var]]) if (var == "depth" ){# pad depth with 0s if within depth_min/max <- r_sp_env[["depth" ]]<- r_env[["depth_min" ]] < p[4 ] & "depth_max" ]] > p[1 ]"depth" ]] <- ifel (is.na (r_depth),ifel (r_ok, 0 , NA ),<- rast (r_sp_env)# show env layers transformed by species preferences plet (y = 1 : nlyr (r_sp_env), col = rev (RColorBrewer:: brewer.pal (11 , "Spectral" )),shared = T, # shared legend collapse = F, tiles = "Esri.OceanBasemap" )<- r_sp# summary(values(r_sp_old, na.rm = T, mat = F)) # Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.0300 0.6750 0.9000 0.7968 1.0000 1.0000 <- app (r_sp_env, fun = mean)# summary(values(r_sp_new, na.rm = T, mat = F)) # Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.5776 0.7355 0.7837 0.7769 0.8000 1.0000 # without ice_con # r_sp_new <- r_sp_env |> # terra::subset( # setdiff( # names(r_sp_env), # "ice_con")) |> # app(fun = mean) # summary(values(r_sp_new, na.rm = T, mat = F)) # Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.4719 0.6694 0.7297 0.7211 0.7500 1.0000 # as product # r_sp_new <- app(r_sp_env, fun = prod) # summary(values(r_sp_new, na.rm = T, mat = F)) # Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.00000 0.00000 0.00000 0.09531 0.00000 1.00000 compare_sp (mapView (r_sp_new - r_sp_old, layer.name = "new - old" )``` ```{r} #| label: ck_prim_prod #| eval: false # https://github.com/marinebon/aquamaps-downscaled/blob/503c62f39552c4ad2417413b13c49d4d4405f3ab/sp-map/functions.R#L92C5-L93C30 # if (b == "PrimProd") # im <- im$multiply(1000) tbl (con_dd, "spp_prefs" ) |> select (prim_prod_max) |> collect () |> pull (prim_prod_max) |> summary ()# Min. 1st Qu. Median Mean 3rd Qu. Max. # 1.62 27.19 43.27 52.55 69.46 253.40 # names(r_h) summary (values (r_h[["prim_prod_mean" ]], na.rm = T, mat = F))# Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.089 1.145 2.668 4.284 5.348 250.926 ``` ```{r} #| label: get sp occurrences #| eval: false # dbListTables(con_dd) # tbl(con_dd, "_tbl_fld_renames") |> # group_by(tbl_old, tbl_new) |> # summarize(n = n(), .groups = "drop") |> # collect() # # tbl_old tbl_new n # <chr> <chr> <dbl> # 1 hspen_r spp_prefs 56 # 2 speciesoccursum_r spp 25 # 3 occurrencecells_r spp_occs 17 # 4 hcaf_r cells 58 # 5 hcaf_species_native spp_cells 7 <- tbl (con_dd, "spp_occs" ) |> filter (sp_key == !! sp_key) |> collect () |> st_as_sf (coords = c ("center_long" , "center_lat" ), remove = F,crs = 4326 )# r_sp_old: -98 , -76.5 , 26 , 30 # r_sp_new: -97 , -80 , 26 , 30 # pts_occs: -97.25, -77.25, 23.25, 30.25 (xmin, xmax, ymin, ymax) # pts_good: -97.25, -77.25, 26.25, 29.25 # pts_ibnd: -97.25, -77.25, 26.25, 29.25 # bb_occs: -97.5 , -77.0 , 26.0 , 29.5 # bb_pad: -98.0, -76.5 , 25.5 , 30.0 |> ext () |> filter (good_cell == 1 ) |> ext () |> filter (in_bound_box == 1 ) |> ext ()# pad <- 0 # bb_occs <- 0.5 # bb_pad <- tbl (con_dd, "spp_occs" ) |> filter (== !! sp_key,== 1 ) |> left_join (tbl (con_dd, "cells" ),by = "cell_id" ) |> summarize (xmin = min (w_limit, na.rm = T) - pad,xmax = max (e_limit, na.rm = T) + pad,ymin = min (s_limit, na.rm = T) - pad,ymax = max (n_limit, na.rm = T) + pad) |> collect () |> pivot_longer (everything ()) |> deframe ()``` - `fao_area_m` : Code number of FAO statistical area to which the cell belongs, for all oceanic and coastal cells. `r # range(values(r_h[["fao_area_m"]], na.rm = T))` [ 18, 88 ] - `fao_area_in` : Code number of FAO statistical area to which the cell belongs, for all inland and coastal cells. `r # range(values(r_h[["fao_area_in"]], na.rm = T))` [ 1, 8 ] ## Get Bio-Oracle v3 higher resolution data - [ Bio-ORACLE : Marine data layers for ecological modelling ](https://www.bio-oracle.org/documentation.php) - [ Bio-ORACLE : Marine data layers for ecological modelling ](https://www.bio-oracle.org/downloads-to-email.php) - [ bio-oracle/biooracler: R package to access Bio-Oracle data via ERDDAP ](https://github.com/bio-oracle/biooracler) - [ ERDDAP - List of All Datasets ](https://erddap.bio-oracle.org/erddap/info/index.html?page=1&itemsPerPage=1000) - [ Copernicus Marine Data Store | Copernicus Marine Service ](https://data.marine.copernicus.eu/products) ```{r} #| label: bio-oracle :: shelf (- oracle/ biooracler,quiet = T)# biooracler:::erddap.bio_oracle.org() # http://erddap.bio-oracle.org/erddap/ :: list_layers () |> :: datatable ()``` ## Match AquaMaps env to Bio-Oracle layers ```{r} :: shelf (quiet = T)|> select (ends_with ("_yn" )) |> names () |> str_replace ("_yn" , "" )# "depth" "temp" "salinity" "prim_prod" "ice_con" "oxy" "land_dist" "extn_rule" ``` - `depth` : terrain_characteristics.bathymetry_min|mean|max## Taxonomic comparisons - Aquamaps: [ Computer Generated Native Distribution Map for _Crepidula depressa_ ](https://aquamaps.org/preMap2.php?cache=1&SpecID=W-Msc-419703) - [ Mapping Parameters ](https://www.aquamaps.org/ShowMapParam.php?SpecID=W-Msc-419703&graph=&user_session=1&bp=-1) - `"W-Msc-419703"` : _Crepidula depressa_ (class = Gastropoda) n_cells = 50```{r} <- "W-Msc-419703" tbl (con_dd, "spp" ) |> filter (sp_key == !! sp_key) |> collect () |> mutate (across (everything (), as.character)) |> pivot_longer (everything ()) |> kable ()``` - [ Crepidula depressa | **SeaLifebase.se** ](https://www.sealifebase.se/summary/Crepidula-depressa.html) - [ **Ecology** ](https://www.sealifebase.se/Ecology/SpeciesEcologySummary.php?StockCode=92041&GenusName=Crepidula&SpeciesName=depressa) - `Substrate` : Benthic: mobile; - `Feeding habit` : filtering plankton - [ **Ecosystems** ](https://www.sealifebase.se/trophiceco/EcosysList.php?ID=151110&GenusName=Crepidula&SpeciesName=depressa) - Ecosystem: Atlantic Ocean, Gulf of Mexico, ...; Type: Sea/Bay/Gulf - [ **Synonyms** ](https://www.sealifebase.se/Nomenclature/SynonymsList.php?ID=151110&SynCode=179777&GenusName=Crepidula&SpeciesName=depressa) [ Crepidula depressa ](https://www.sealifebase.se/Nomenclature/SynonymSummary.php?ID=179777&CAS_SPC=&Status=accepted%20name&Synonymy=senior%20synonym&Combination=original%20combination&GenusName=Crepidula&SpeciesName=depressa&SpecCode=151110&SynonymsRef=83435&Author=Say,%201822&Misspelling=0) | Say, 1822 | accepted name | Yes | senior synonym | original combination- [ WoRMS - World Register of Marine Species - Crepidula depressa Say, 1822 ](https://www.marinespecies.org/aphia.php?p=taxdetails&id=419703) - unaccepted: [ WoRMS - World Register of Marine Species - Crepidula depressa Deshayes, 1830 ](https://www.marinespecies.org/aphia.php?p=taxdetails&id=572470) - [ Crepidula depressa - Wikipedia ](https://en.wikipedia.org/wiki/Crepidula_depressa) - [ Search "Crepidula depressa" | GBIF.org ](https://www.gbif.org/search?q=Crepidula%20depressa) - [ Crepidula depressa Say, 1822 ](https://www.gbif.org/species/5856498) - [ Crepidula depressa Say, 1822 ](https://www.gbif.org/species/155138324) \ - [ Crepidula depressa Deshayes, 1830 ](https://www.gbif.org/species/8275070) - [ Crepidula depressa Say, 1822 - OBIS.org ](https://obis.org/taxon/419703)