erDiagram
%% https://mermaid.js.org/syntax/entityRelationshipDiagram.html
%% tables
sdm_datasets {
str ds_key PK "dataset key"
str name_short
str name_full
str link
arr links_other
str source_broad
str source_detail
arr regions
str description
str citation
str response_type
str taxa_groups
int year_pub
date date_obs_beg "date of observations start"
date date_obs_end "date of observations end (not inclusive)"
date date_env_beg "date of environmental data start"
date date_env_end "date of environmental data end (not inclusive)"
}
sdm_models {
str ds_key PK "dataset key"
int mdl_id PK "model ID"
str sp_key FK
str population
str time_interval
str var "variable, e.g.: cv, n, se"
str description}
sdm_species {
str ds_key PK "dataset key"
str sp_key PK "species key"
int aphia_id FK "singular taxa ID (shared parent if multiple)"
arr aphia_ids FK "array of all taxa IDs (if multiple)"
str sp_scientific
str sp_common
str description }
sdm_geometries {
str ds_key PK "dataset key"
int geom_id PK "geometry ID"
geo geom "geometry"}
sdm_values {
str ds_key PK "dataset key"
int mdl_id PK "model ID"
int geom_id PK "geometry ID"
dbl val "value"}
taxa {
int aphia_id PK "unique ID at MarineSpecies.org"
str scientific_name
str common_name
str rank
str iucn_cat "IUCN Red List extinction risk category"
}
%% relationships
sdm_datasets ||--|{ sdm_models : ds_key
sdm_datasets ||--|{ sdm_species : ds_key
sdm_datasets ||--|{ sdm_geometries : ds_key
sdm_models ||--|{ sdm_species : "ds_key,sp_key"
sdm_geometries ||--|{ sdm_values : "ds_key,geom_id"
sdm_models ||--|{ sdm_values : "ds_key,mdl_id"
taxa ||--|{ sdm_species : aphia_id
taxa }o--|{ sdm_species : aphia_ids
Create SDM Tables
storing species distribution model outputs in a database for analysis
1 Species Distribution Models
The distribution of models varies geographically by extent, spatial analytical units, taxa, time period and output variable. To store the original data, we ingest data into the following database public schema:
Specific columns:
- sdm_datasets:
name_short
short name in form of {source_broad} {regions} {taxa_groups} {response_type}name_long
original nameresponse_typeone of: occurrence, range, suitability, probability or densitytaxa_groups
one or more of: fish, invertebrates, marine mammals, cetaceans, sea turtles, seabirds, etc.
- sdm_models`
time_interval
time interval in format of {start}/{duration} per ISO 8601, e.g. “2019-02/P1M” is a duration of one month starting February, 2019. See https://en.wikipedia.org/wiki/ISO_8601#Time_intervals.
1.1 Data Dictionary
Data types: - str
string: variable-length character string, up to 1 GB; - ser
serial: 1 to 2,147,483,647; - bser
bigserial: 1 to 9,223,372,036,854,775,807; - int
integer: -2,147,483,648 to 2,147,483,647; - dbl
double precision: 15 decimal digits of precision;
2 Database Table and Column Naming Conventions
- Table names are plural and use all lower case.
- Unique identifiers are suffixed with:
*_idfor unique integer keys;*_keyfor unique string keys;*_seqfor auto-incrementing sequence integer keys.
- Column names are singular and use snake_case.
- Foreign keys are named with the singular form of the table they reference, followed by _id.
- Primary keys are named id.
3 CREATE TABLE SQL
Using the raw Mermaid diagram from above, ChatGPT 4 generated the SQL to (re-)create the tables, relationships and column comments. See chat: SQL Tables & Relationships.
In pgAdmin, selected public schema and ran the following SQL:
-- Drop existing tables in reverse order of dependency
DROP TABLE IF EXISTS sdm_values CASCADE;
DROP TABLE IF EXISTS sdm_geometries CASCADE;
DROP TABLE IF EXISTS sdm_models CASCADE;
DROP TABLE IF EXISTS sdm_species CASCADE;
DROP TABLE IF EXISTS sdm_datasets CASCADE;
DROP TABLE IF EXISTS taxa CASCADE;
-- Recreate tables
CREATE TABLE sdm_datasets (
ds_key VARCHAR PRIMARY KEY,
name_short VARCHAR,
name_full VARCHAR,
link VARCHAR,
links_other VARCHAR[], -- Adjust according to actual data type
source_broad VARCHAR,
source_detail VARCHAR,
regions VARCHAR[], -- Adjust according to actual data type
description TEXT,
citation TEXT,
response_type VARCHAR, -- Add constraint or check if needed
taxa_groups VARCHAR,
year_pub INT,
date_obs_beg DATE,
date_obs_end DATE,
date_env_beg DATE,
date_env_end DATE
);
CREATE TABLE taxa (
aphia_id INT PRIMARY KEY,
scientific_name VARCHAR,
common_name VARCHAR,
rank VARCHAR,
iucn_cat VARCHAR
);
CREATE TABLE sdm_species (
ds_key VARCHAR,
sp_key VARCHAR,
aphia_id INT,
aphia_ids INT[],
sp_scientific VARCHAR,
sp_common VARCHAR,
description TEXT,
PRIMARY KEY (ds_key, sp_key),
FOREIGN KEY (ds_key) REFERENCES sdm_datasets(ds_key) ON DELETE CASCADE,
FOREIGN KEY (aphia_id) REFERENCES taxa(aphia_id) ON DELETE SET NULL
);
CREATE TABLE sdm_models (
ds_key VARCHAR,
mdl_id INT,
sp_key VARCHAR,
population VARCHAR,
time_interval VARCHAR,
var VARCHAR,
description TEXT,
PRIMARY KEY (ds_key, mdl_id),
FOREIGN KEY (ds_key) REFERENCES sdm_datasets(ds_key) ON DELETE CASCADE,
FOREIGN KEY (sp_key, ds_key) REFERENCES sdm_species(sp_key, ds_key) ON DELETE CASCADE
);
CREATE TABLE sdm_geometries (
ds_key VARCHAR,
geom_id INT,
geom GEOMETRY,
PRIMARY KEY (ds_key, geom_id),
FOREIGN KEY (ds_key) REFERENCES sdm_datasets(ds_key) ON DELETE CASCADE
);
CREATE TABLE sdm_values (
ds_key VARCHAR,
mdl_id INT,
geom_id INT,
val DOUBLE PRECISION,
PRIMARY KEY (ds_key, mdl_id, geom_id),
FOREIGN KEY (ds_key, mdl_id) REFERENCES sdm_models(ds_key, mdl_id) ON DELETE CASCADE,
FOREIGN KEY (ds_key, geom_id) REFERENCES sdm_geometries(ds_key, geom_id) ON DELETE CASCADE
);
-- Add comments to columns
COMMENT ON COLUMN sdm_datasets.name_short IS 'short name in form of {source_broad} {regions} {taxa_groups} {response_type}';
COMMENT ON COLUMN sdm_datasets.name_full IS 'original name';
COMMENT ON COLUMN sdm_datasets.response_type IS 'one of: occurrence, range, suitability, probability or density';
COMMENT ON COLUMN sdm_datasets.taxa_groups IS 'one or more of: fish, invertebrates, marine mammals, cetaceans, sea turtles, seabirds, etc.';
COMMENT ON COLUMN sdm_datasets.date_obs_beg IS 'date of observations start';
COMMENT ON COLUMN sdm_datasets.date_obs_end IS 'date of observations end (not inclusive)';
COMMENT ON COLUMN sdm_datasets.date_env_beg IS 'date of environmental data start';
COMMENT ON COLUMN sdm_datasets.date_env_end IS 'date of environmental data end (not inclusive)';
COMMENT ON COLUMN sdm_models.var IS 'variable, e.g.: cv, n, se';
COMMENT ON COLUMN sdm_models.time_interval IS 'time interval in format of {start}/{duration} per ISO 8601, e.g. "2019-02/P1M" is a duration of one month starting February, 2019. See https://en.wikipedia.org/wiki/ISO_8601#Time_intervals.'
is a valid ISO 8601 interval notation. This format combines a specific starting period (in this case, a month) with a duration. Here's how to interpret it:
2019-02: Indicates the start of the interval, which is February 2019.
';
COMMENT ON COLUMN taxa.iucn_cat IS 'IUCN Red List extinction risk category';4 ALTER TABLE SQL
Including a placeholder here for subsequent modifications, liked adding columns to the tables.
4.1 Update geom to show on tile server
To show on tile.marinesensitivity.org.
ALTER TABLE public.sdm_geometries ALTER COLUMN geom TYPE geometry (MultiPolygon, 4326);5 Function to Fetch SDM with spatial data
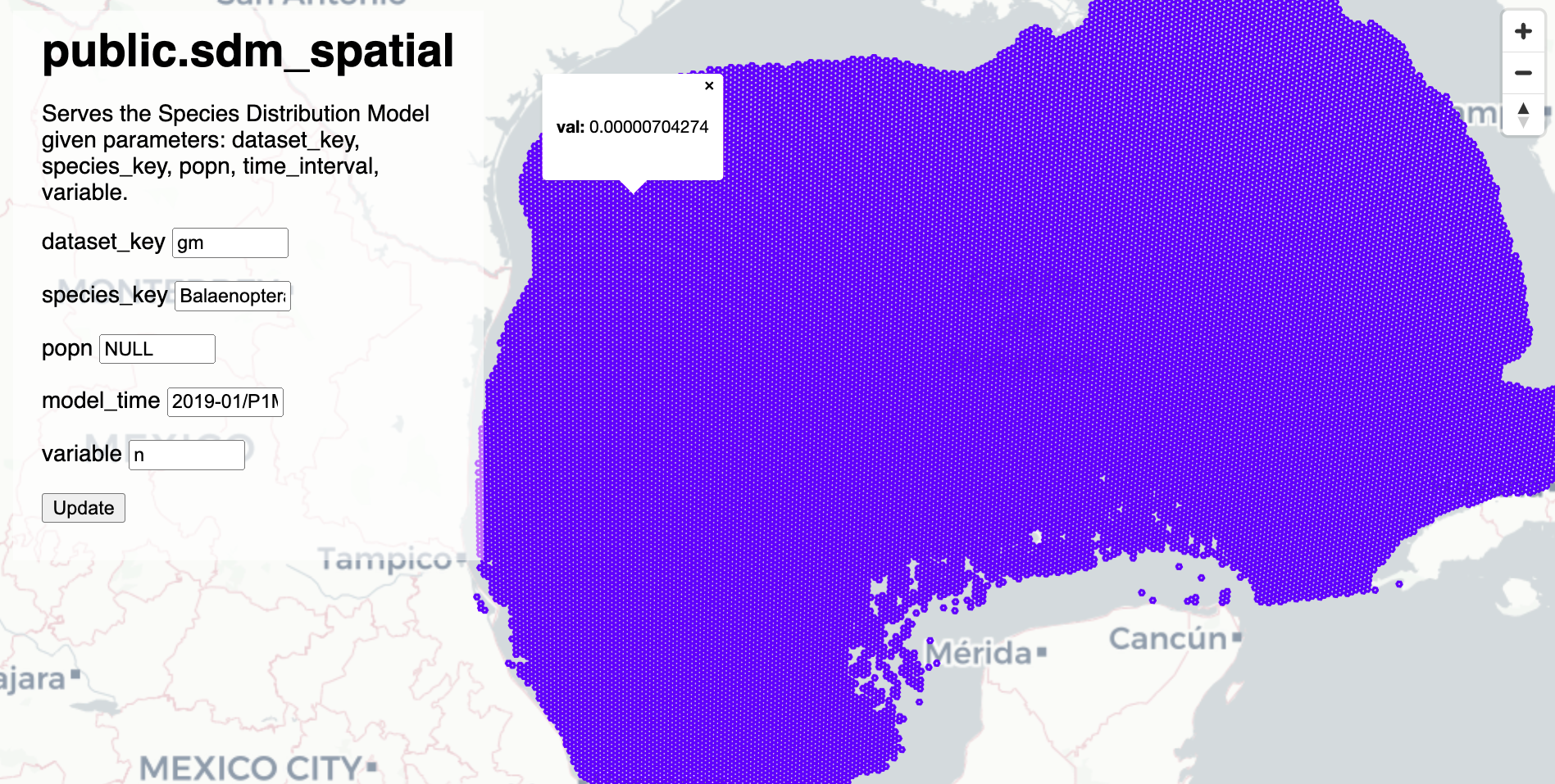
Try resulting function:
-- DROP FUNCTION sdm_spatial(integer,integer,integer,text,text,text,text,text);
CREATE OR REPLACE
FUNCTION public.sdm_spatial(
z integer, x integer, y integer,
dataset_key text default 'gm',
species_key text default 'Balaenoptera ricei',
popn text default 'NULL',
model_time text default '2019-01/P1M',
variable text default 'n')
RETURNS bytea
AS $$
DECLARE
result bytea;
BEGIN
WITH
bounds AS (
SELECT ST_TileEnvelope(z, x, y) AS geom ),
m AS (
SELECT v.val, g.geom
FROM (
(SELECT ds_key, mdl_id
FROM public.sdm_models
WHERE
ds_key = dataset_key AND
sp_key = species_key AND
-- population = NULLIF(popn, 'NULL') AND
CASE WHEN popn = 'NULL' THEN population IS NULL ELSE population = popn END AND
time_interval = model_time AND
var = variable ) AS d
INNER JOIN public.sdm_values AS v ON (
d.ds_key = v.ds_key AND
d.mdl_id = v.mdl_id )
INNER JOIN public.sdm_geometries AS g ON (
d.ds_key = g.ds_key AND
v.geom_id = g.geom_id ) )
WHERE val IS NOT NULL ),
mvtgeom AS (
SELECT ST_AsMVTGeom(ST_Transform(m.geom, 3857), bounds.geom) AS geom, m.val
FROM m, bounds
WHERE ST_Intersects(m.geom, ST_Transform(bounds.geom, 4326)) )
SELECT ST_AsMVT(mvtgeom, 'default')
INTO result
FROM mvtgeom;
RETURN result;
END;
$$
LANGUAGE 'plpgsql'
STABLE
PARALLEL SAFE;
COMMENT ON FUNCTION public.sdm_spatial IS 'Serves the Species Distribution Model given parameters: dataset_key, species_key, popn, time_interval, variable.';